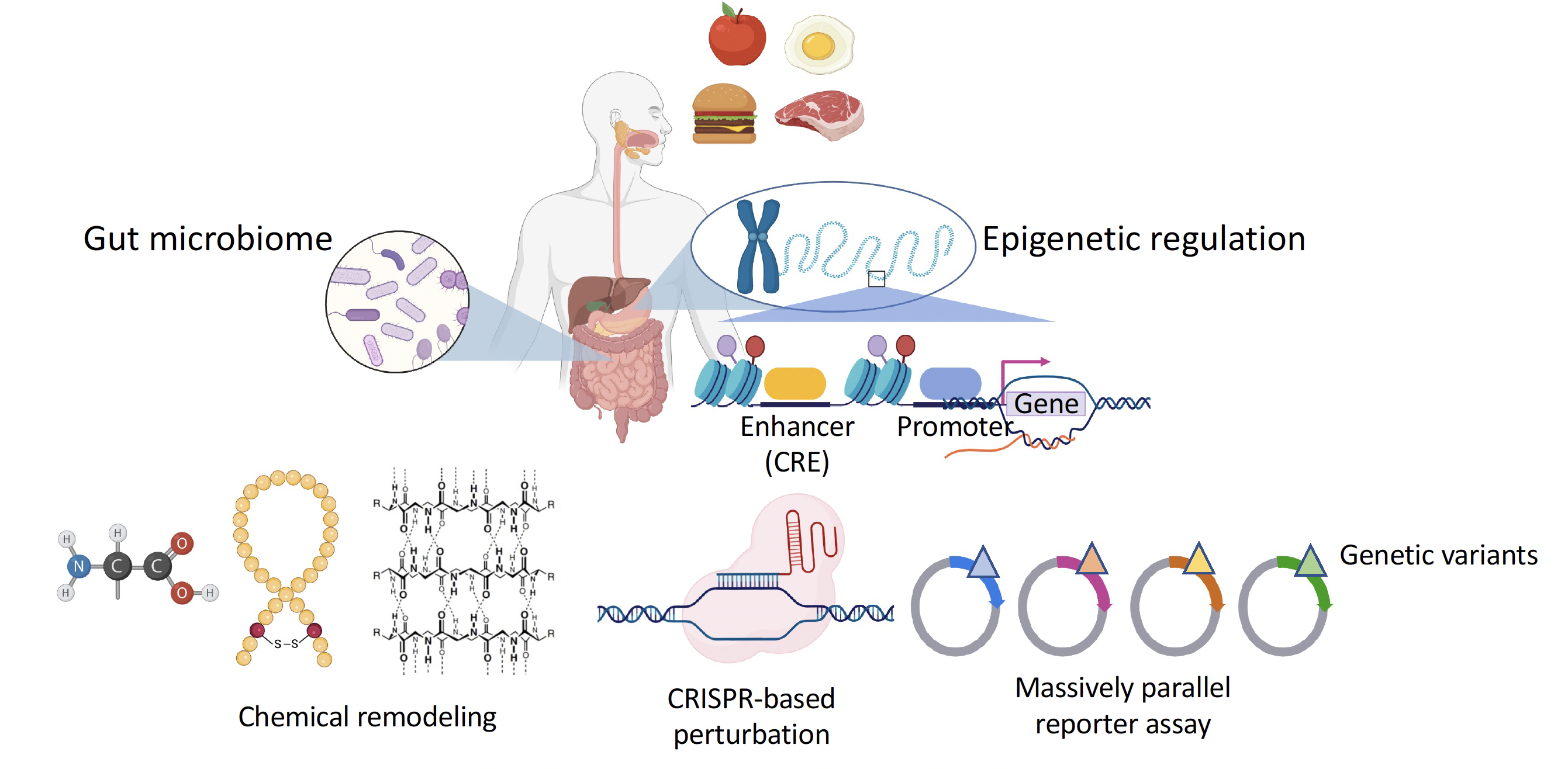
We develop and utilize high-throughput functional assays to understand how gene regulatory programme is controlled under different conditions. We are currently focusing on the following questions:
- How does the human genome respond to environmental stimuli?
- How does the dysbiosis in the gut microbiome lead to the development of liver diseases?
- How does the gut microbiota affect colorectal cancer development?